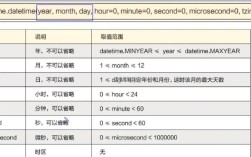
Of course! Curve fitting in Python is a common task used in science, engineering, finance, and data analysis to find a mathematical function that best represents a set of data points.
Here's a comprehensive guide covering the most popular and effective methods, from simple to advanced.
The Core Idea
Given a set of data points (x, y), we want to find the parameters of a function f(x, p) (where p is a vector of parameters) that minimizes the difference between the observed y values and the values predicted by the function, f(x, p).
The most common measure of this difference is the Sum of Squared Errors (SSE):
SSE = Σ(y_i - f(x_i, p))^2
We will explore three main approaches:
- Polynomial Fitting (Simple and built-in)
- Non-Linear Least Squares (The standard, powerful method using
scipy.optimize.curve_fit) - Interpolation (For finding a function that passes exactly through all points)
Setup: Necessary Libraries
First, make sure you have the essential libraries installed. scipy is the key one for advanced fitting.
pip install numpy matplotlib scipy
Now, let's import them in Python:
import numpy as np import matplotlib.pyplot as plt from scipy.optimize import curve_fit from scipy.interpolate import interp1d
Method 1: Polynomial Fitting (numpy.polyfit)
This is the simplest method for fitting a polynomial of a specified degree to your data. It's fast and works well when you suspect the relationship is polynomial.
How it works:
numpy.polyfit(x, y, deg) performs a least-squares polynomial fit. It returns a vector of coefficients p where p[0]*x**deg + p[1]*x**(deg-1) + ... + p[deg] is the fitted polynomial.
Example: Fitting a 2nd-degree polynomial (a parabola)
# 1. Generate some sample data with noise
np.random.seed(42)
x_data = np.linspace(0, 10, 20)
y_data = 2.5 * x_data**2 - 3.0 * x_data + 1.5 + np.random.normal(0, 8, len(x_data))
# 2. Fit a 2nd-degree polynomial
# polyfit returns the coefficients [a, b, c] for ax^2 + bx + c
coefficients = np.polyfit(x_data, y_data, 2)
print("Coefficients (a, b, c):", coefficients)
# 3. Create the polynomial function from the coefficients
poly_function = np.poly1d(coefficients)
# 4. Generate points for the smooth fitted curve
x_fit = np.linspace(0, 10, 200)
y_fit = poly_function(x_fit)
# 5. Plot the results
plt.figure(figsize=(8, 6))
plt.scatter(x_data, y_data, label='Original Noisy Data', color='red')
plt.plot(x_fit, y_fit, label='Fitted Polynomial', color='blue', linewidth=2)'Polynomial Curve Fitting')
plt.xlabel('X-axis')
plt.ylabel('Y-axis')
plt.legend()
plt.grid(True)
plt.show()
Pros:
- Extremely simple to use.
- Very fast.
Cons:
- Limited to polynomial functions.
- Choosing the right degree (
deg) is crucial and can be tricky. A high degree can lead to overfitting.
Method 2: Non-Linear Least Squares (scipy.optimize.curve_fit)
This is the most powerful and flexible method. It can fit any function you can define to your data, as long as it's non-linear in its parameters. This is the go-to tool for most scientific curve fitting.
How it works:
You must define the function you want to fit, providing an initial guess for its parameters. curve_fit then adjusts these parameters to minimize the sum of the squared residuals.
Example: Fitting an exponential decay curve
Let's say we have data that we believe follows the form y = a * exp(-b * x) + c.
# 1. Define the function to fit.
# The first argument must be x.
# The following arguments are the parameters to be fitted (a, b, c).
def exp_decay_func(x, a, b, c):
return a * np.exp(-b * x) + c
# 2. Generate sample data based on this function with some noise
np.random.seed(42)
x_data = np.linspace(0, 4, 50)
y_true = 10 * np.exp(-1.5 * x_data) + 2
y_data = y_true + np.random.normal(0, 0.8, len(x_data))
# 3. Provide an initial guess for the parameters (a, b, c)
# This is important! A good guess helps the algorithm converge.
initial_guess = (8, 1, 1) # Guess for a, b, c
# 4. Perform the curve fit
# popt: Optimal parameters (fitted values for a, b, c)
# pcov: Covariance matrix of the parameters (useful for error estimation)
popt, pcov = curve_fit(exp_decay_func, x_data, y_data, p0=initial_guess)
print("Fitted parameters (a, b, c):", popt)
# 5. Generate points for the smooth fitted curve
x_fit = np.linspace(0, 4, 200)
y_fit = exp_decay_func(x_fit, *popt) # The * unpacks the popt tuple
# 6. Plot the results
plt.figure(figsize=(8, 6))
plt.scatter(x_data, y_data, label='Original Noisy Data', color='red')
plt.plot(x_fit, y_fit, label='Fitted Exponential', color='green', linewidth=2)'Non-Linear Curve Fitting')
plt.xlabel('X-axis')
plt.ylabel('Y-axis')
plt.legend()
plt.grid(True)
plt.show()
Key Points for curve_fit:
- Function Definition: Your function must take
xas its first argument. - Initial Guess (
p0): Providing a reasonable initial guess is critical. If the guess is too far from the true values, the algorithm may fail to find the solution. - Bounds: You can constrain parameters to be within a certain range using the
boundsargument.# Example: forcing parameter 'a' to be positive lower_bounds = [0, -np.inf, -np.inf] upper_bounds = [np.inf, np.inf, np.inf] popt, pcov = curve_fit(exp_decay_func, x_data, y_data, p0=initial_guess, bounds=(lower_bounds, upper_bounds))
Method 3: Interpolation (scipy.interpolate)
Sometimes you don't want to model the underlying trend; you just want a smooth curve that passes exactly through all your data points. This is called interpolation.
How it works:
Interpolation constructs a new function that "connects the dots." It's not suitable for noisy data but is great for plotting a smooth line through a sparse set of precise measurements.
Example: Using a cubic spline interpolator
# 1. Generate some clean, sparse data
x_data = np.linspace(0, 10, 11)
y_data = np.sin(x_data) + np.random.uniform(-0.1, 0.1, len(x_data)) # Add tiny bit of noise for realism
# 2. Create the interpolation function
# 'kind' can be 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic'
f_interp = interp1d(x_data, y_data, kind='cubic')
# 3. Generate points for the smooth interpolated curve
x_fit = np.linspace(0, 10, 200)
y_fit = f_interp(x_fit)
# 4. Plot the results
plt.figure(figsize=(8, 6))
plt.scatter(x_data, y_data, label='Original Data', color='black', zorder=5)
plt.plot(x_fit, y_fit, label='Cubic Spline Interpolation', color='purple', linewidth=2)'Interpolation')
plt.xlabel('X-axis')
plt.ylabel('Y-axis')
plt.legend()
plt.grid(True)
plt.show()
Pros:
- Passes through all data points.
- Excellent for visualizing data without assuming a model.
Cons:
- DO NOT USE for noisy data. It will fit the noise, leading to a very wiggly curve that doesn't represent the true trend (this is called overfitting).
- Extrapolation (predicting outside the range of your data) is very unreliable.
Summary and When to Use Each Method
| Method | Library | Use Case | Key Features |
|---|---|---|---|
| Polynomial Fitting | numpy |
Simple, suspected polynomial relationship. | Easy, fast. Limited |