Of course! This is an excellent question because joblib and pandas are two fundamental libraries in the Python data science ecosystem, and they are often used together.
Here's a comprehensive breakdown of their relationship, how they work together, and best practices.
The Core Relationship: Data Structures vs. Computation
- Pandas: Is the library for data manipulation and analysis. Its primary data structures are the
DataFrame(a 2D table, like a spreadsheet) and theSeries(a 1D array, like a column). Pandas excels at loading, cleaning, transforming, and exploring data. - Joblib: Is a library for pipelining Python functions and persisting objects. Its primary use cases are:
- Caching: Speeding up computations by storing the results of long-running functions.
- Persistence (Serialization): Saving Python objects to disk and loading them back into memory.
In short: Pandas creates/manages your data. Joblib helps you save/load that data and speed up the code that processes it.
The Most Common Use Case: Saving and Loading Pandas Objects
This is the most frequent interaction between the two libraries. While Pandas has its own to_pickle() and read_pickle() functions, joblib is often preferred for larger datasets and certain object types.
Why use joblib over Pandas' pickle?
- Memory Efficiency:
joblibis optimized for large NumPy arrays, which are the building blocks of Pandas DataFrames. It can save arrays in a compressed, memory-mapped format, which is much more efficient for large datasets. - Better Handling of Complex Objects: It can sometimes handle more complex Python objects nested within a DataFrame more robustly than the standard
picklelibrary.
How to Save a DataFrame with joblib
You use joblib.dump().

import pandas as pd
import joblib
import numpy as np
# 1. Create a sample DataFrame
data = {
'feature1': np.random.rand(1000000),
'feature2': np.random.randn(1000000),
'target': np.random.randint(0, 2, 1000000)
}
df = pd.DataFrame(data)
print("DataFrame created:")
print(df.head())
# 2. Save the DataFrame to a file
# The '.joblib' extension is a common convention
joblib.dump(df, 'my_dataframe.joblib')
print("\nDataFrame saved to 'my_dataframe.joblib'")
How to Load a DataFrame with joblib
You use joblib.load().
# 3. Load the DataFrame from the file
loaded_df = joblib.load('my_dataframe.joblib')
print("\nDataFrame loaded from file:")
print(loaded_df.head())
# 4. Verify that the loaded DataFrame is identical to the original
print(f"\nOriginal and loaded DataFrames are equal: {df.equals(loaded_df)}")
Compression with joblib
A key advantage of joblib is the ability to compress the saved file, which is crucial for saving disk space.
# Save with compression (e.g., using 'zlib' or 'gzip')
joblib.dump(df, 'my_dataframe_compressed.joblib', compress=3)
# Load the compressed file
loaded_compressed_df = joblib.load('my_dataframe_compressed.joblib')
print("\nLoaded from compressed file. Is it equal?", df.equals(loaded_compressed_df))
The compress parameter (from 0 to 9, where 9 is maximum compression) can dramatically reduce file size at the cost of a slightly longer save time.
Advanced Use Case: Caching for Performance
joblib's second main feature is caching. This is incredibly useful when you have a function that takes a long time to run, especially one that operates on Pandas DataFrames.
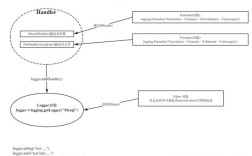
The joblib.Memory object creates a "cache" directory. When you decorate a function with memory.cache, the first time it's called, the result is computed and saved to the cache. On all subsequent calls with the exact same arguments, the result is loaded from the cache instead of being recomputed.
Example: Caching a Data Processing Function
Let's say we have a slow data cleaning function.
import pandas as pd
import numpy as np
import time
from joblib import Memory
# 1. Set up a memory cache
# This will create a 'joblib_cache' directory
memory = Memory(location='./joblib_cache', verbose=0)
# 2. Create a large DataFrame to process
raw_data = {
'A': np.random.rand(100000),
'B': np.random.randn(100000),
'C': np.random.choice(['foo', 'bar', 'baz'], 100000)
}
raw_df = pd.DataFrame(raw_data)
# 3. Define a slow function that processes the DataFrame
@memory.cache
def slow_data_processing(df):
"""
A function that simulates a slow computation on a DataFrame.
"""
print("Performing slow data processing...")
time.sleep(5) # Simulate a 5-second task
# Some actual processing
processed_df = df.copy()
processed_df['A_squared'] = processed_df['A'] ** 2
processed_df['B_log'] = np.log(np.abs(processed_df['B']) + 1e-6) # Avoid log(0)
# Groupby operation
summary = processed_df.groupby('C').agg({
'A_squared': 'mean',
'B_log': 'std'
}).rename(columns={
'A_squared': 'mean_A_squared',
'B_log': 'std_B_log'
})
return processed_df, summary
# --- First Run ---
print("--- First Run: Cache will be populated ---")
start_time = time.time()
processed_df, summary = slow_data_processing(raw_df)
end_time = time.time()
print(f"First run took: {end_time - start_time:.2f} seconds")
print("\nSummary DataFrame:")
print(summary)
# --- Second Run ---
print("\n--- Second Run: Loading from cache ---")
start_time = time.time()
# The function will be called, but the result is loaded from cache instantly
processed_df_cached, summary_cached = slow_data_processing(raw_df)
end_time = time.time()
print(f"Second run took: {end_time - start_time:.4f} seconds")
print("\nCached Summary DataFrame:")
print(summary_cached)
# Verify they are the same
print("\nAre the summaries identical?", summary.equals(summary_cached))
When you run this, you will see the "Performing slow data processing..." message only once. The second run will be nearly instantaneous.
How NOT to Use Them: joblib.Parallel with Pandas
A common point of confusion is using joblib.Parallel to parallelize operations on a Pandas DataFrame. This is almost always the wrong approach.
joblib.Parallel is designed to execute independent Python functions in parallel. Pandas operations, however, are highly optimized and often already parallelized under the hood (e.g., using NumPy, which can use multiple cores).
Bad Practice (Using joblib.Parallel on a DataFrame):
from joblib import Parallel, delayed
# A simple function to apply to a row
def process_row(row):
# This is a trivial operation, but imagine it's complex
return row['A'] * row['B']
# Using Parallel to apply this to each row is very inefficient
# compared to Pandas' built-in vectorized operations.
# results = Parallel(n_jobs=2)(delayed(process_row)(row) for _, row in df.iterrows())
Why is this bad?
- Overhead: Spawning processes/threads has overhead. For simple operations, this overhead is greater than the benefit of parallelization.
df.iterrows()is slow: This is one of the slowest ways to iterate over a DataFrame.- Ignores Pandas' Vectorization: Pandas is built on NumPy, which performs operations on entire arrays at once (vectorization), which is much faster than row-by-row Python loops.
Good Practice (Using Pandas' Built-in Methods):
Pandas provides highly optimized, often parallel, methods for common operations.
-
Vectorized Operations: Use built-in arithmetic.
# This is extremely fast and uses optimized C/Python code. df['C'] = df['A'] * df['B']
-
df.apply(): While still a loop,df.apply()is much faster thaniterrows()and can be parallelized using its ownengine='numba'or by usingswifterlibrary.# This is better than a manual loop, but still slower than vectorization df['C'] = df.apply(lambda row: row['A'] * row['B'], axis=1)
-
df.groupby().apply(): For group-wise operations, this is the idiomatic way. You can passn_jobsto some versions ofgroupbyto parallelize the group